diff options
| author | ROUVREAU Vincent <vincent.rouvreau@inria.fr> | 2020-05-31 11:01:24 +0200 |
|---|---|---|
| committer | ROUVREAU Vincent <vincent.rouvreau@inria.fr> | 2020-05-31 11:01:24 +0200 |
| commit | 5141caeff03f1e8c8c4ccae1ee7ca43fbcb2925f (patch) | |
| tree | 7123e5daffca4fc3307e49453e403a143eb06b02 | |
| parent | 3b91ae863718c402a991c5807579f578d266a04b (diff) | |
It was not markdown but rst...
| -rw-r--r-- | src/python/CMakeLists.txt | 2 | ||||
| -rw-r--r-- | src/python/introduction.rst (renamed from src/python/introduction.md) | 15 | ||||
| -rw-r--r-- | src/python/setup.py.in | 2 |
3 files changed, 11 insertions, 8 deletions
diff --git a/src/python/CMakeLists.txt b/src/python/CMakeLists.txt index d4cb7477..fee6b6f5 100644 --- a/src/python/CMakeLists.txt +++ b/src/python/CMakeLists.txt @@ -236,7 +236,7 @@ if(PYTHONINTERP_FOUND) file(COPY "gudhi/weighted_rips_complex.py" DESTINATION "${CMAKE_CURRENT_BINARY_DIR}/gudhi") # Other .py files - file(COPY "introduction.md" DESTINATION "${CMAKE_CURRENT_BINARY_DIR}/") + file(COPY "introduction.rst" DESTINATION "${CMAKE_CURRENT_BINARY_DIR}/") add_custom_command( OUTPUT gudhi.so diff --git a/src/python/introduction.md b/src/python/introduction.rst index 3277e0ac..2cc4642e 100644 --- a/src/python/introduction.md +++ b/src/python/introduction.rst @@ -1,6 +1,11 @@ -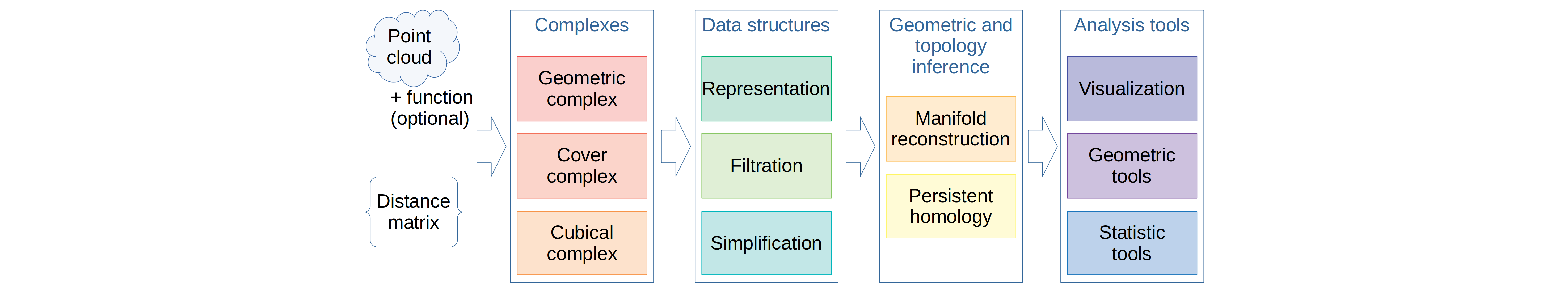 +.. figure:: + https://gudhi.inria.fr/images/gudhi_architecture.png + :figclass: align-center + :width: 100 % + :alt: GUDHI -#Introduction +Introduction +============ The Gudhi library is an open source library for Computational Topology and Topological Data Analysis (TDA). It offers state-of-the-art algorithms @@ -9,6 +14,7 @@ represent them, and algorithms to compute geometric approximations of shapes and persistent homology. The GUDHI library offers the following interoperable modules: + * Complexes: * Cubical * Simplicial: Rips, Witness, Alpha and Čech complexes @@ -24,7 +30,4 @@ The GUDHI library offers the following interoperable modules: * Persistence diagram and barcode For more information about Topological Data Analysis and its workflow, please -refer to the [Wikipedia TDA dedicated page][1]. - - [1]: https://en.wikipedia.org/wiki/Topological_data_analysis - +refer to the `Wikipedia TDA dedicated page <https://en.wikipedia.org/wiki/Topological_data_analysis>`_. diff --git a/src/python/setup.py.in b/src/python/setup.py.in index ff000a2c..4725bb44 100644 --- a/src/python/setup.py.in +++ b/src/python/setup.py.in @@ -65,7 +65,7 @@ for module in pybind11_modules: # read the contents of introduction.md this_directory = path.abspath(path.dirname(__file__)) -with open(path.join(this_directory, 'introduction.md'), encoding='utf-8') as f: +with open(path.join(this_directory, 'introduction.rst'), encoding='utf-8') as f: long_description = f.read() setup( |
